a <- c(1, 2, 3, 4)
plot(a)4 Getting to know R and RStudio
In Chapter 2, we went through all the trouble of installing and setting up the tools needed to become data scientists. It is now assumed that everything was indeed installed and working. In this chapter we will introduce the usage of R and Rstudio. First we will set up and customize RStudio and then learn how to communicate with R.
4.1 The Anatomy of RStudio
The appearance of RStudio can be changed for a more pleasant user experience. I like a dark theme as it is easier on the eye. We can also move the different components of RStudio. I like to have the console on the top right and the source on the top left. I think this makes it easier to see output when coding interactively. All this will be clearer as thing evolve, but for now, start R Studio, go to Tools > Global options and make it personal (see Figure 4.1)!
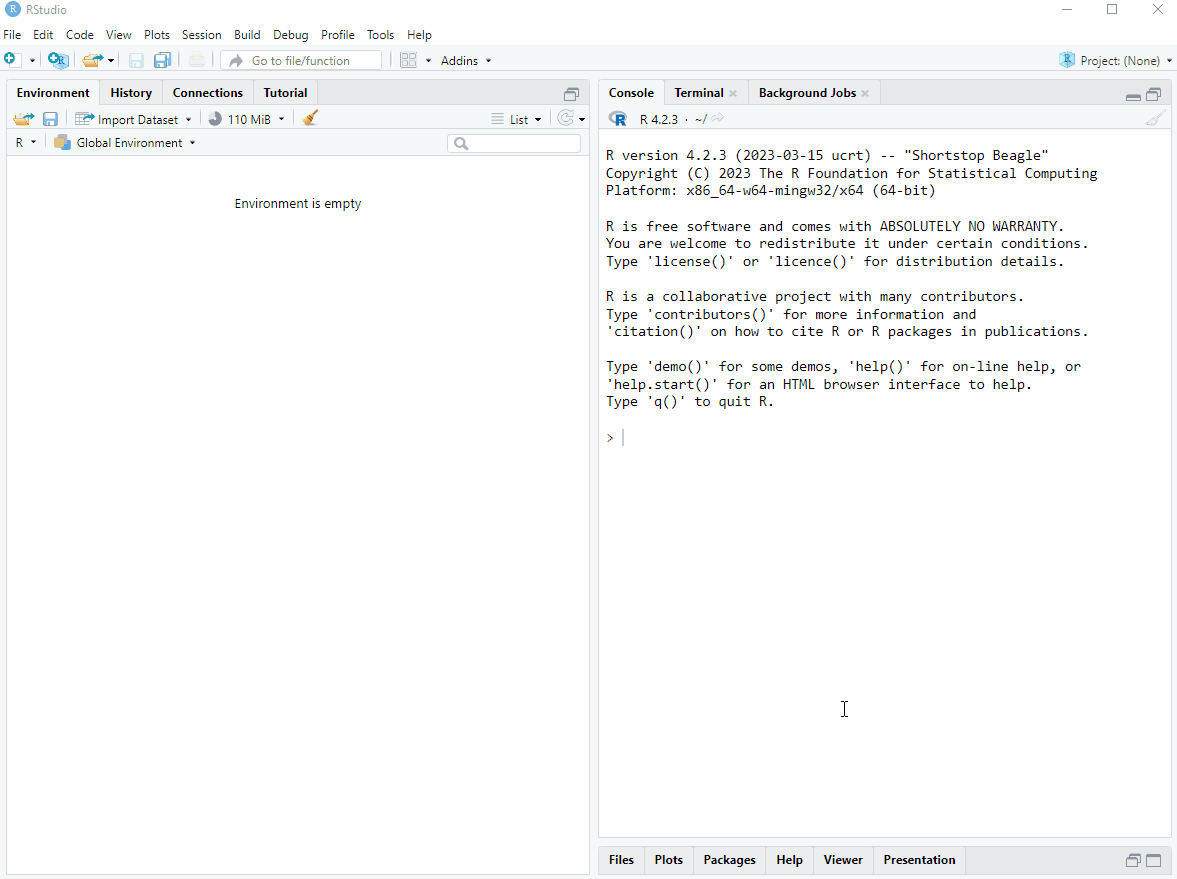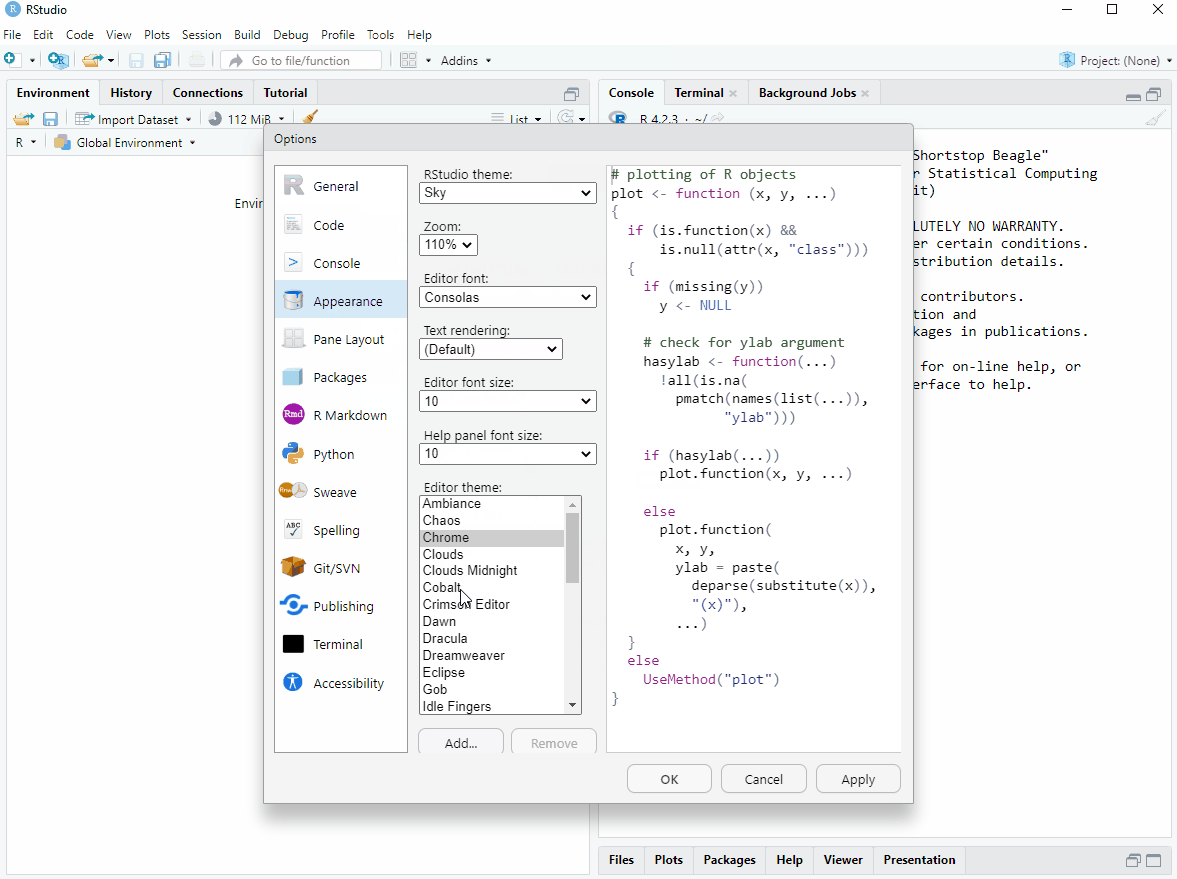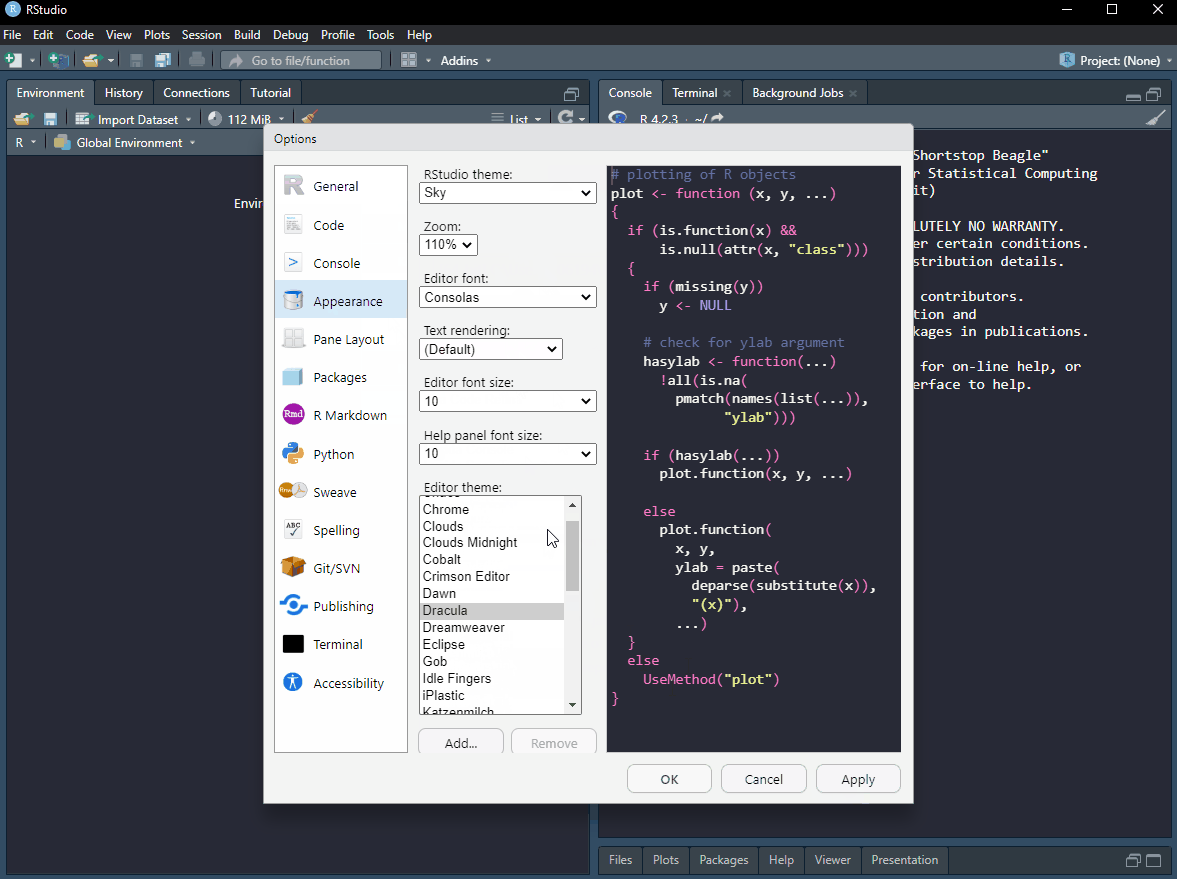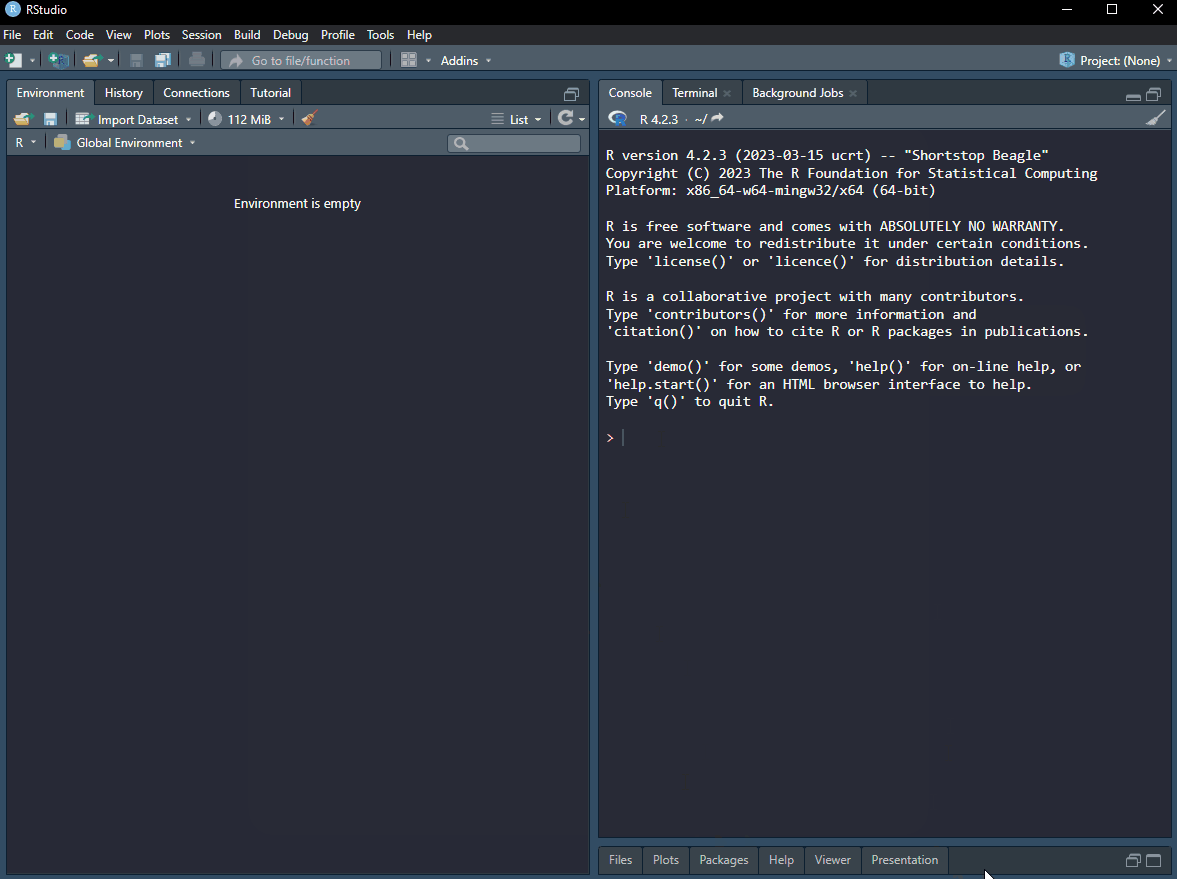
As you may have spotted in the image above, it is possible to change the font of your editor. I like Fira code.
Source editor: Is where scripts are edited.
Environment: In R, the environment is where data variables and structures are saved during execution of code.
Script: Your script is the document containing your computer code. This is your computer program (using a loose definition of a software program).
Variables: In R, variables are containers for data values.
Workspace: This is your environments as represented on your computer. A workspace can be, but should not be saved between sessions.
4.1.1 The source editor
The source editor is where you edit your code. When writing your code in a text-file, you can call it a script, this is essentially a computer program where you tell R what to do. It is executed from top to bottom. You can send one line of code, multiple lines or whole sections into R. In the image below (Figure 4.2), the source window is in the top left corner.
4.1.2 Environment
The environment is where all your objects are located. Objects can be variables or data sets that you are working with. In RStudio the environment is listed under the environment tab (bottom left in the image).
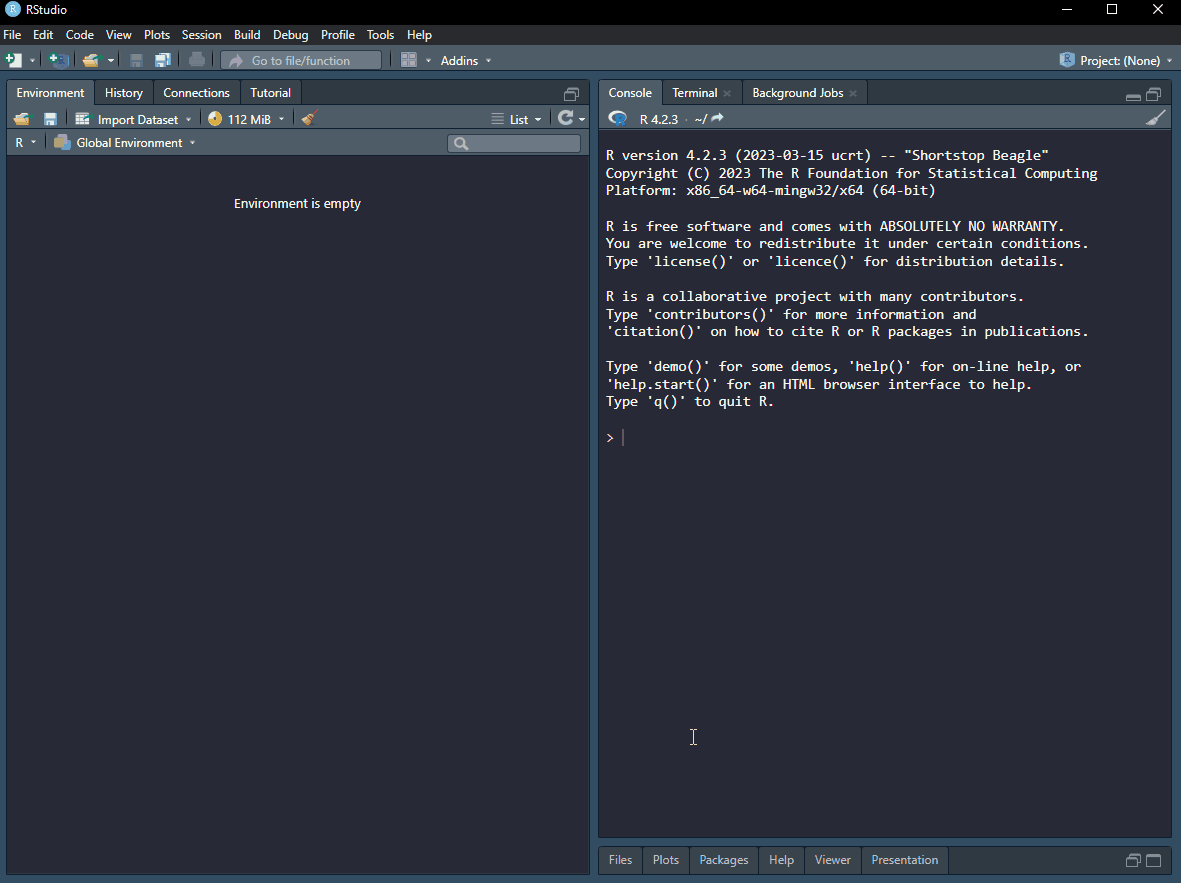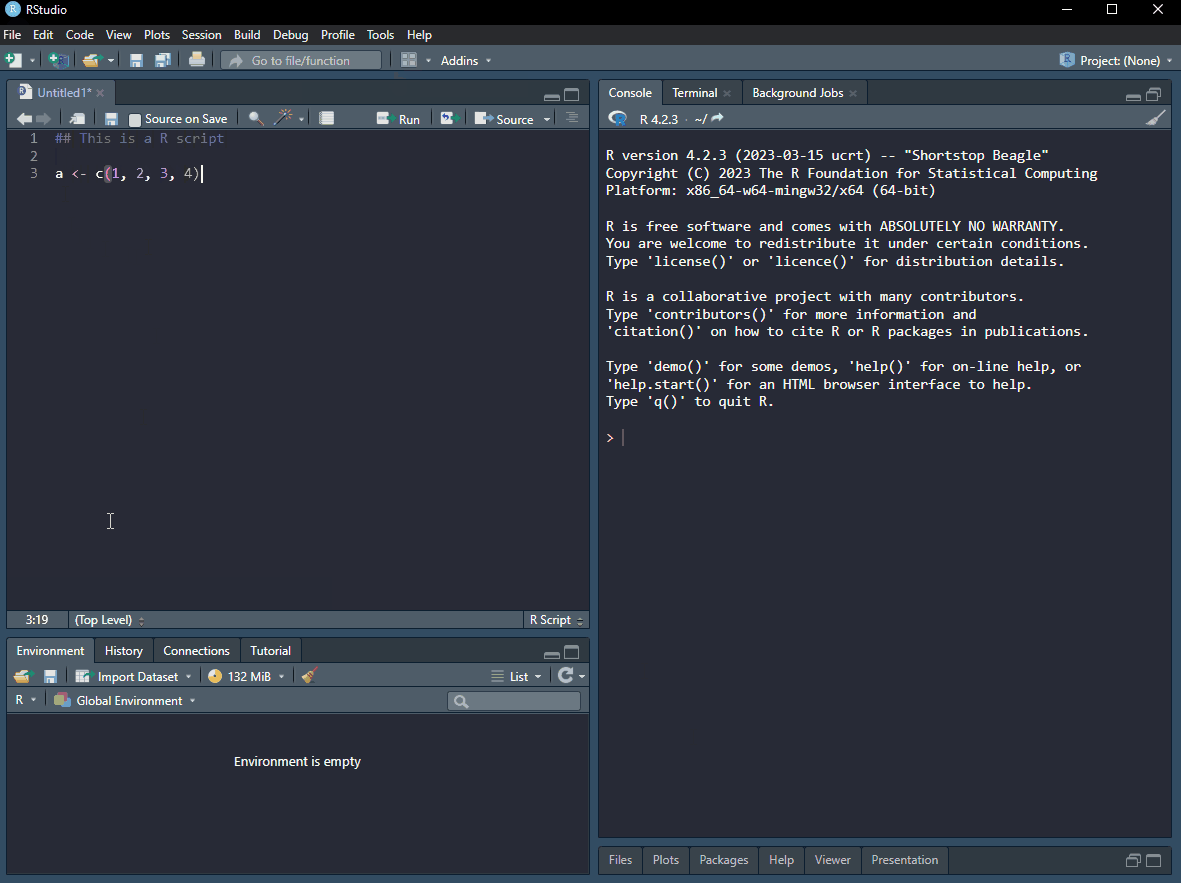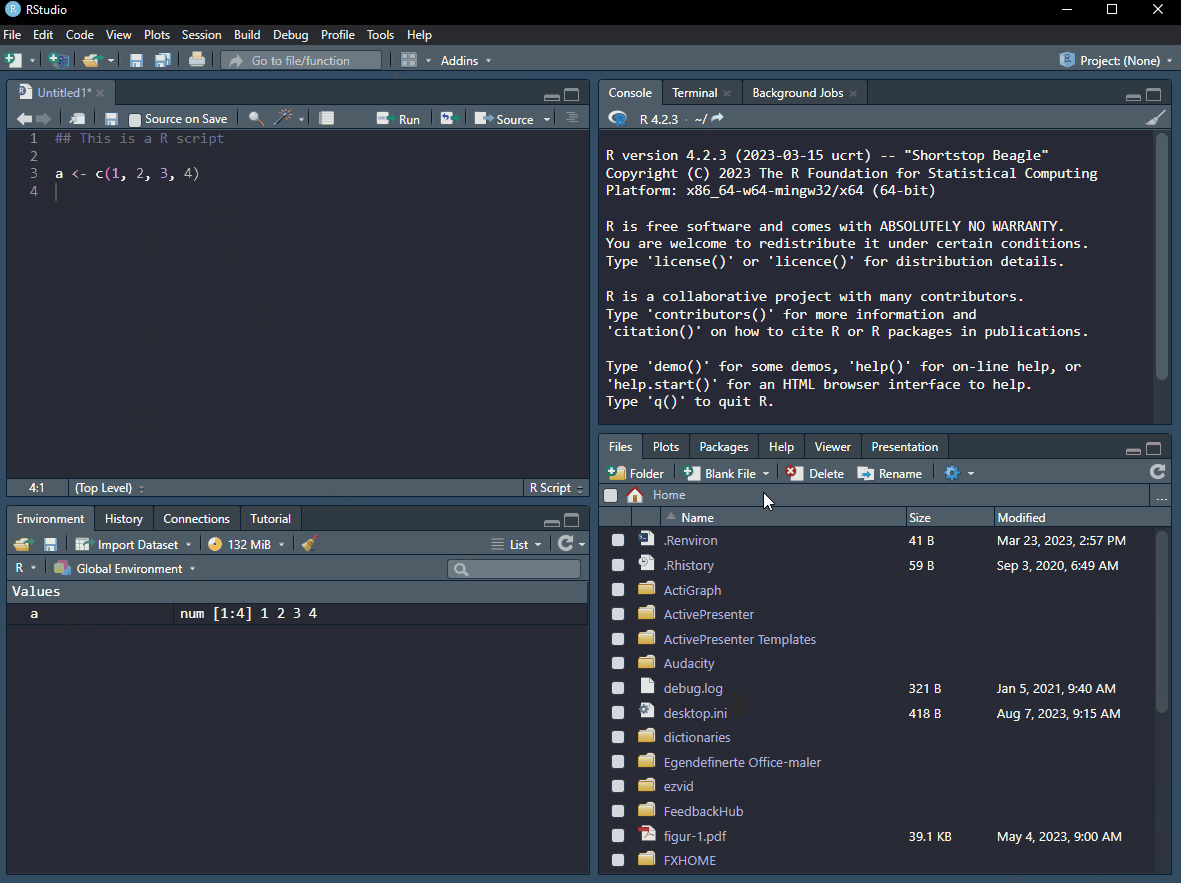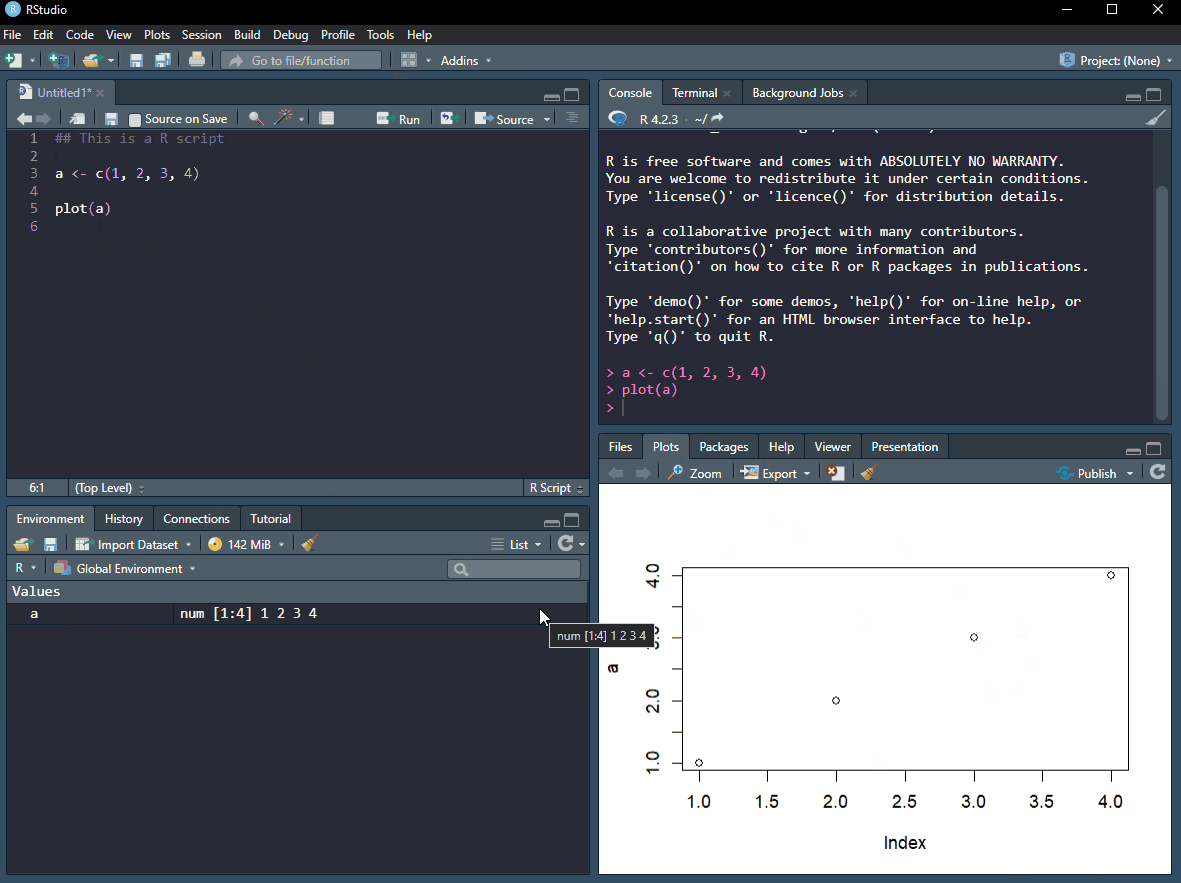
Copy the code below to a R script. To run it line by line, set your cursor on the first line a press Ctrl+Enter. What happened in your environment? Press Ctrl+Enter again and you will see a plot in the plot window. Amazing stuff!
4.1.3 The console
By pressing Ctrl+Enter from the script, as described above, you sent your code to the console. You can also interact with R directly here. By writing a in the console and hitting enter you will get the value from the object called a. This means that it is also where output from R is usually printed. In the image below, the console is in the top right corner.
4.1.4 Files, plots, packages and help files
In RStudio files are accessible from the Files tab. The files tab shows the files in you root folder. The root folder is where R will search for files if you tell it to. We will talk more about the root folder later in connection with projects. Plots are displayed in the Plot tab. Packages are listed in the packages tab. If you access the help files, these will be displayed in the help tab. In the image below all these tabs are in the bottom right corner. More on help files and packages later.
4.2 Reproducible data science using RStudio
When starting to work more systematically in RStudio we will set some rules that will allow for reproducible programming. Remember from Chapter 2 that part of a fully reproducible study is software/code that produces the results. It turns out that when working interactively with R you can fool yourself to believe that you have included all steps needed to produce some results in your script. However, variables may be stored in your environment but not by assigning values to them in your script. This will become a problem if you want to share your code, a certain value/variable needed to make the program work may be missing from your script.
To avoid making such a mistake it is good practice not to save variables in your environment between sessions, everything should be scripted and documented and assumed not defined elsewhere. In RStudio we can make an explicit setting Not to save the workspace (See ?fig-saveworkspace).
4.3 Basic R programming, first steps
You have already been asked to run commands in your version of RStudio, below we will run some very basic commands here on this website to get first hand experience with the R language. Below you will find several script boxes that you may edit before pressing Run Code. When you run code, the results you would see in your terminal will appear below the script.
4.3.1 Objects and assignment
Everything in R are objects (Chambers 2009). What does this mean? Let’s say that we want to store some information in R, this information is a value, let’s say the number 12. To store this information we will assign the value to an object and call this object twelve. An object is a container of data “of all kinds” (Chambers 2009, 111) that we create in our working memory to hold our data (the value). We assign the value to our object by using an assignment operator. The most common assignment operator is <-. Think of the assignment operator as an arrow pointing from the value towards the object, like this:
twevle <- 12We can also reverse the direction of the assignment operator and make the object and value change places.
12 -> twelveTo call an object, or tell R that we want to look at our object, we would simply type the object name in our R console. We may try this in the R script box below. I have already entered some information in the script, these are comments starting with the hash symbol (#). The comments tells us what to do. You can enter R code on the line below the comments, R will ignore everything following a comment sign and start to interpret code again on the subsequent line. When you have entered your code, press “Run Code” to inspect the results.
It is also possible to use the equal sign (=) to assign data to objects. The equal sign used as an assignment operator, as in twelve = 12, is equivalent to twelve <- 12.
4.3.2 R as a giant calculator
R can make use of all basic arithmetic operators like plus (+), minus (-), divided by (*) and multiplied by (/). These operators can be used on objects, and directly on values. In the script box below, try to calculate (and store) the following:
\[y = 5 * 2 + 1 * 0.5\] Then use the object and add 10, store the new result in a new object called z.
A possible solution to the above challenge would be
y <- 5 * 2 + 1 * 0.5
z <- y + 10The result of your computations should be 20.5. In the above example we have discovered that mathematical expression can be written using values and objects already stored in the environment.
We can also use functions to perform mathematical operations on objects. Examples of such functions are log(), exp(), abs() and sqrt(). A function is a special object that itself can be use other objects (or variables/values) as input. A function often takes arguments, these arguments are supplied to the function inside its brackets.
The log() function returns the natural logarithm of a numeric object. Mathematically, the log function returns the exponent (\(x\)) with the base \(e\) that give us our input number \(y\) (\(e^x = y\)). To get the natural logarithm of 100 we would use R and write log(100) which results in 4.6051702. This corresponds to \(100 = 2.718282 ^ {4.60517}\). The base \(e\) must be raised to the power of 4.60517 to yield 100.
What is the natural logarithm of 50? Try to calculate it in the script box below.
We can of course make similar computations on stored objects.
Above we have learned that numbers and objects that store numbers can be used in computations using basic arithmetic operators and functions that perform mathematical operations.
4.3.3 Different types of data
Above we have worked on numeric data. These are values represented either as integers (whole numbers; e.g. 1, 2, 3) or what is sometimes called “double” or “numerical”, i.e. decimal numbers (e.g. 1.2 or 2.781).
In the code example below we will store an integer and a double and inspect its “class”, the class decides what R can do with an object.
num <- as.numeric(2.456)
int <- as.integer(2)
class(num)
class(int)Use the script box below to test out the code example. What is the purpose of the class function?
There are other types of data that we can use in R, these are character, logical and complex (we will not discuss complex numbers further here).
A character or string value can be though of as text, e.g. "this is a character" could be the data contained in a character object. A logical value is either TRUE or FALSE. This type of data is also known as Boolean
As mentioned above, the type of data restricts what operations that can be performed using the data. For example, we cannot do mathematical operations on characters or logicals. Try out the code below and inspect the results. What does the error message tells you?
We can define data types by using functions such as as.numeric() or as.character() these will tell R to try to convert data to a specific type. If this is not possible you will get an error message. Try the code below and inspect the error message.
The result of the conversion of a character to a numeric is NA, that can be read as Not Available. NA is an example of a protected symbol in R. We cannot use this as a name of an object. It is used to indicate missingness, or missing values.
In the above section we have identified different type of data that are used in R
4.3.4 Combining data
So far we have work on single valued objects. This is not very efficient. Conveniently, R has an efficient way of working with data using vectors. A vector is a structure for combining data of the same type. For example, we can construct a numeric vector of heights using the combine function c(). Let’s say height <- c(1.74, 1.81, 1.51, 1.92). An additional vector of weights can also be constructed as weight <- c(85.1, 81.1, 48.9, 88.4). We can use these vector to do calculations, these calculations will be “vectorized”.
The body mass index is calculated as
\[BMI = \frac{\text{Weight (kg)}}{\text{Height (m)}^2}\] Using vectorized operations we can calculate BMI for each row of the two vectors defined above simply using BMI <- weight/height^2. Modify the code below to inspect each vector and the resulting BMI vector.
Vectors can be combined into data frames. These are convenient tabular two-dimensional representations of multiple, equal length vectors. To combine the vectors defined above into a data frame we could directly put them in a data frame. In the example below we use df to name the data frame. To add a new variable (or vector) to the data frame we can use the $ operator which creates a new variable (or overwrites it!). Notice also that we access weight and height in the existing data.frame using the $ operator.
Alternatively we may access specific rows and columns of data frames using brackets. The syntax is df[row,column]. To access all row of a specific column, for example “weight” we would write df[,1]since weight is the first column of the data frame called “df”.
Try to calculate BMI using the row index method explained above.
A data frame can combine different data types in the same data structure. The data are, as indicate above related by row as one row contains e.g. weight and height from one individual. We might add information on the individuals by adding variables of different types. Modify and run the code below to inspect the data frame.
We can further combine data into a list. A list can contain different data structures or values/objects. A list can even contain other lists. To combine objects into a list we simply put them into the list() function. To access objects in lists we can use double brackets. E.g., using the code below we could access the second item in the list using my_list[[2]]. Objects in lists can also be named, in such cases we can use the $ operator to access them. Modify the code below to explore this concept.
4.3.5 Logical operations and conditions
In the future you will select observations based on some specific conditions. This could for example mean that you would want to keep all observations where the variable X is greater than 5. To communicate this to R we would create a vector of TRUE and FALSE. R will keep all observations that satisfy our condition and therefore are TRUE.
In the script box below, we first construct a vector of numbers followed by a logical test. The test will result in TRUE when the condition is satisfied. Notice that the “test” gives you a vector of TRUE/FALSE.
In the example above we used the ‘greater than’ operator (>). There are a few more usefull operators:
| Operator | Meaning |
|---|---|
== |
equal to |
!= |
not equal to |
< |
less than |
> |
greater than |
<= |
less than or equal to |
>= |
greater than or equal to |
Try to modify the script box above to test out the different operators.
Using the “AND” operator (&) we can add conditions that needs to be fulfilled to produce TRUE. This might be useful when two or more conditions needs to be satisfied. In addition to our values stored in my_values in the script box above, we might want to see the condition colors == "green" satisfied as well, where colors is a vector of colours. Run the codse below and inspect the results
We can store the results in a vector or use it to select values in a vector. Using brackets on a vector (or data frame) we can select observations based on a logical vector (produced by logical tests).
Using the “NOT” operator (!) we can perform negation on any of the logical operators. Let’s say we want have all observations that do not satisfy our filter above. Run the code below and inspect the results. Notice the parentheses which indicates that we put the negation on the whole expression.
The “OR” operator (|) gives us the possibility to select values that satisfies one or both of tqo conditions.
my_values > 5 | colors == "green"
Try to put the above in square brackets and filter one of the vectors to see that you get what you anticipate.
Finally, we could test if a logical vector contains TRUE or if all values are TRUE. To do this we would use the functions any() and all().
4.3.6 Functions
We mentioned above that everything in R is an object. This is true even when we talk about functions. Functions are special kind of objects, they contain code that upon execution perform certain tasks. A function may need to have certain arguments specified. Arguments are user input into the function to specify how the functions should behave. A common usage of a function is to do something with data that you, the user, supply to the function.
Let’s specify a function to see what it does. We have a vector of numbers, let’s say my_values. We want to construct a function that calculates the mean of that vector. The function may later be used to calculate averages of other vectors so we should try to make it as general as possible. Let’s start with the design of the actual calculation1.
1 Don’t be afraid of mathematics! Take it slow and translate it to your language. Some books on statistics are a lot easier to read if you are prepared to read simple mathematical expressions. Sometimes it is also good to be able to write an expression. Mathematics, like computer code, is also a language. To learn a new language we need to try not to be afraid!
The mean (\(\bar{x}\)) of a vector (\(x_i\)) is calculated as
\[\begin{align} \bar{x}&=\frac {1}{n} \sum_{i=1}^{n}x_{i} \\ &= \frac {x_{1}+x_{2}+\cdots +x_{n}}{n} \end{align}\]
Which we can read as “one or the number of observations times the sum of all observations”. Or, as in the second row, the sum of all observations divided by the number of observations.
We can translate this to code. A simple way to do this is to use other functions, in this case length() that returns the number of observations (or length) in a vector, and sum() that gives us the sum of all observations in a numeric vector. In the code block below I have simply combined data with the expression needed to calculate the average.
some_values <- c(3, 4, 5, 6, 7, 8)
1/length(some_values) * sum(some_values)The next step is to put the code into a function. A function is defined using a special function, called function! Confusing? Yes. Let’s see how it is done.
my_mean_function <- function(DATA) {
1/length(DATA) * sum(DATA)
}Using function we define the function called my_mean_function. Using the assignment operator it is stored in our environment (R’s working memory). The function “body” contains the code. It says that it will use an object called DATA that should be given as an argument in the function call, notice that we have defined the function with an argument called DATA.
Since we are not storing the output from the calculation 1/length(DATA) * sum(DATA) in any new object inside the function running the function in our R session will return the mean of what ever we define as DATA.
In the script block below we have everything defined. Try out the function by modifying the code so that it prints my_mean.
Of course, there is already a function that will give you the mean of a numeric vector and it is called mean(). In the simple case, the mean() function takes one argument x that should be a numeric vector. It could look like this:
mean(my_values) # Calculate the mean of your vector.Defining functions for yourself can be a very efficient way of performing data analysis, but most functions that you need are already specified in R. Other people have already gone trough the trouble of defining functions that are easy to use for specific tasks.
4.3.7 Functions and packages
The R ecosystem consists of packages. These are collections of functions organized in a systematic manner. Functions are created to perform a specialized task. And packages often have many function used to do e.g. analyses of a specific kind of data, or more general task such as making figures or handle data.
In this course we will use many different packages, for example dplyr, tidyr and ggplot2. dplyr and tidyr are packages used to transform and clean data. ggplot2 is used for making figures.
To install a package, you use the install.packages() function. You only need to do this once on your computer (unless you re-install R). You can write the following code in your console to install dplyr.
install.packages("dplyr")Alternatively, click “Packages” and “Install” and search for the package you want to install. To use a package, you have to load it into your environment. Use the library() function to load a package.
library("dplyr")We will become familiar with packages as we move along in the course.
4.4 Basics R programming: Installing and using swirl
Swirl is a great way to get to know how to talk with R. Swirl consists of lessons that run in your R console where you interactively will be able to learn basic concepts. Start RStudio and install swirl by typing the following into your console:
install.packages("swirl")When swirlis installed you will need to load the package This means that all functions that are included in package becomes available to you in your R session. To load the package you use the library function.
library("swirl")When you run the above command in your console you will get a message saying to call swirl() when you are ready to learn. At this stage, run the course “R Programming: The basics of programming in R”. Swirl will ask if you want to install it. After installation, just follow the instructions in the console. To get out of swirl, just press ESC.
4.5 File formats for editing and executiong R code
A confusing part of using R is that we are really just able to communicate with R through the console. There are however a lot of methods to do this, and save your input, and output for later. This is central to how we will work with R: We create some input (code), R returns results, like numbers, text or figures and these can be formatted to be saved in different formats.
4.5.1 R scripts
The most basic file format for R code is an R script, as we have already touched upon. An R script contains code and comments. Code is executed by R and comments are ignored. Ideally, R scripts are commented to improve readability of what the do. Commenting code is also a good way of creating a roadmap of what you want to do. In the image below (Figure 4.3), R code is written based on a plan written with comments. Note that when a line starts with at least one # it is interpreted by R as a comment.
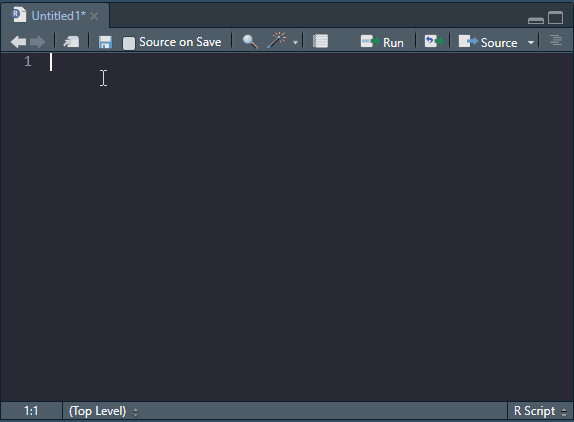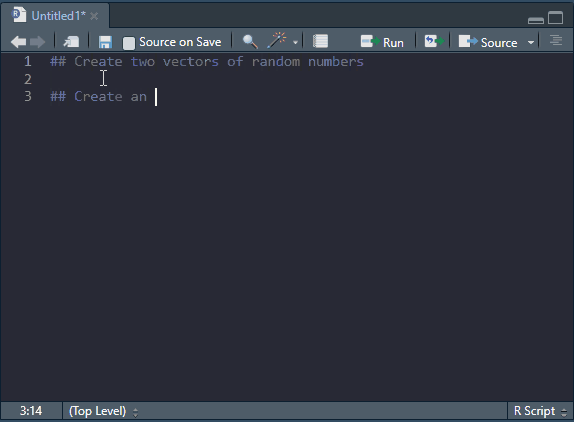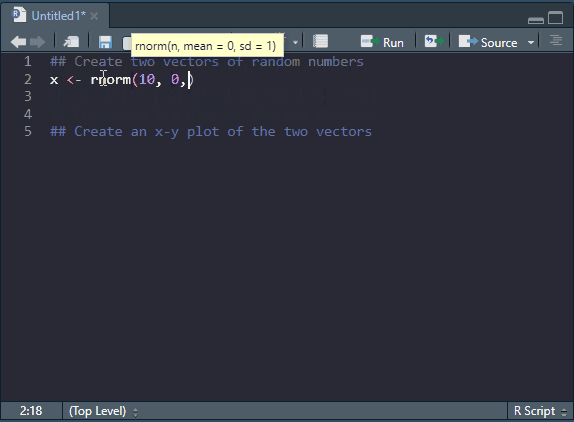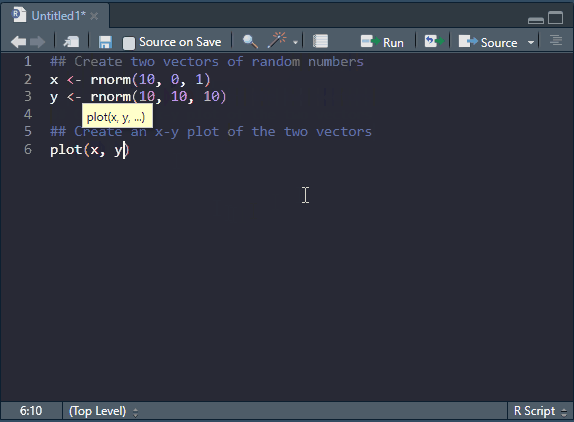
Try the code for yourself to see what it produces. The details will be covered later.
## Create two vectors of random numbers
x <- rnorm(10, 0, 1)
y <- rnorm(10, 10, 10)
## Create an x-y plot of the two vectors
plot(x, y)In RStudio code will be highlighted with different colours to indicate e.g. functions and and arguments in functions. RStudio has the capabilities to do this for multiple languages.
4.5.2 R markdown and quarto files
The more advanced file formats for R are RMarkdown (.rmd) and quarto (.qmd) files. These have the capabilities of combining formatted text with computer code. The source document may contain multiple pieces of code organized in code chunks together with text formatted with markdown syntax. A meta data field in the top of the source file specifies settings for the conversion to output formats. Multiple output formats are available, including HTML, word and PDF. The image below shows the basic outline of a very simple quarto file destined to create a HTML document.
Notice also that RStudio offers an visual editor where the output is approximated and formatting is available from a menu.
Adding headlines and makes it possible to navigate the document through the outline or the list of components in the bottom of the document.
R markdown and quarto have many similarities as the basic organization is similar between the two. The text parts are written using a special syntax, markdown. The point of markdown is that you will use the same syntax that is later possible to convert to multiple formats. The syntax let’s you do all formatting explicitly, for example instead of getting your mouse to superscript some text you can add syntax a^2^ to achieve a2.
A full guide to RMarkdown can be found on the official R markdown web pages. I suggest you take the time to get an overview of this language as it will make you more fluent in the tools that enables reproducible computing. When writing R markdown, it is handy to have a cheat sheet close by when writing, here is an example for Rmarkdown, and here is another one for quarto 2.
2 Cheat sheets are available in R Studio: Help > Cheatsheets
We will cover the quarto publication system in more details in later chapters.